Synapse List for
from
-
Synapses are grouped into three tables,
one for each synapse type:
- Gap junctions
- Presynaptic: synapses where -- is presynaptic
- Postsynaptic: synapses where -- is postsynaptic
-
Each table is further organized by the synaptic partners.
There are two types of rows:
- Summary rows: corresponds to a partner(s); shows the total number and sections of synapses with that partner
- Individual rows: corresponds to a synapse; shows the database/series (redundant), section number/z-coordinate (of the middle of the synapse), and synapse ID (=contin number; it's also a link to the Synapse (EM) Viewer).
- Click on Synapse ID to see EM (opens new tab).
- Bracketed Cells (e.g.[PVX]) denotes an inferred process identification (not traced to Cell Body)
- unk denotes an unknown neurite process identification
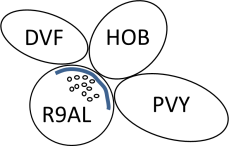
- In synapse lists, the listed order of postsynaptic cells in polyads represents the clockwise order of the cells around the presynaptic density, electron micrographs viewed looking toward the head. Thus, R9AL->DVF,HOB,PVY represents a synapse that appears like the diagram below in the electron micrograph.
- A 'nmj_' in front a synapse denotes a neuromuscular junction.
- Occasionally, synapes do not display properly in the maps. In cases where there is a discrepancy between the maps and this synapse list, this synapse list should be considered correct.
- Link to this page, directly to this cell: ?db=--&cell=--&listtype=synapse
Partner List for
--
from
--
- This is essentially a summary version of Synapse List, where we group synapses by partner (ignores polyadic structure; see below on exactly how).
-
We have 4 tables: 1 for electrical,
and 3 for chemical: 'pre', 'post', 'post-post'.
Say the chosen cell is 'X'.
The 'pre' table shows the postsynaptic partners
when 'X' is presynaptic.
The 'post' table shows the presynaptic partner
when 'X' is postsynaptic.
The 'post-post' table shows the postsynaptic partners
when 'X' is also postsynaptic.
A synapse may count towards several tables; for example, a chemical synapse of the form 'X -> X,X,Y,X' would contribute to all three tables. More precisely, here is how many times the syanpse contributes to each relevant row- 'pre', Partner='X': 3 times
- 'pre', Partner='Y': 1 time
- 'post', Partner='X': 1 time
- 'post', Partner='Y': 0 times
- 'post-post', Partner='X': 2 times
- 'post-post', Partner='Y': 1 time
- Bracketed Cells (e.g.[PVX]) denotes an inferred process identification (not traced to Cell Body)
- unk denotes an unkown neurite process identification
- Link to this page, directly to this cell: ?db=--&cell=--&listtype=partner